UT’s Non-Digital Biodiversity Specimens Join the Global Digital Revolution
The prestigious journal BioScience just released "Natural History Collections: Advancing the Frontiers of Science," a compilation of recent natural history collection-related papers that sheds light on the importance of digitizing and publishing collections data, and the substantial obstacles confronting collections staff working on that. All of us curators in the Biodiversity Center know these obstacles all too well, having been laboring on them for decades. As Dr. Alex Wild (Curator, Entomology Collections) pointed out, we're making progress despite major limitations, and his collection now being 1% digitized, though perhaps sounding trivial to some, is indeed a major accomplishment.
Here, I'll provide a broader perspective, exploring all of our collections combined, since, at long last, I finally can! All of our data from all four primary Biodiversity Center collections are now easily explored anywhere in the world via a single online portal - the international GBIF.org (Global Biodiversity Information Facility).
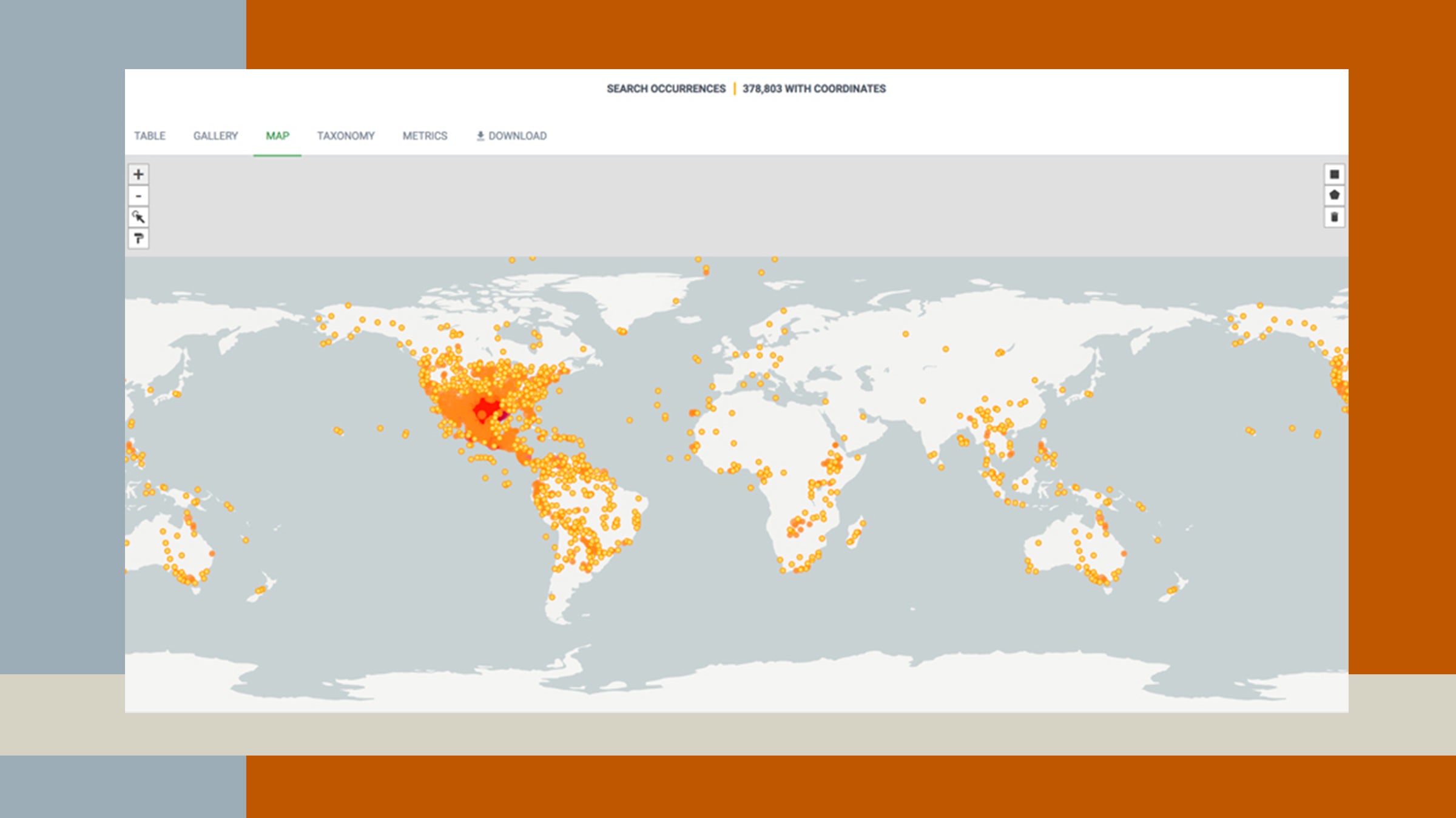
So, let's take a tour. This map in GBIF (also above) of our combined data quickly reveals the near global origin of our 843,803 specimen occurrence records, and the hotspot on Texas and vicinity illustrates our focus. GBIF's taxonomy tab points out that plants comprise 57% of our total specimens, and that in the animal world we have 327,483 records of vertebrates (Fishes 213,656, Amphibians 59,190, Reptiles 54,312) and 22,797 Arthropods (a part of invertebrates). One can dive deeper with dynamic data summaries revealing how many records we have at any taxonomic level (e.g. class, order, family, or species), or do the same for other categories (geographic, temporal, etc.). The metrics page reveals (for any subset of the data) the temporal distribution of specimen collection dates, and has summaries of the (relatively minor) data quality issues we continue to work on fixing. We currently have images of 7200 of our specimens explorable in GBIF's gallery view. Soon, all other collections’ datasets will join the Fish collection’s data in iDigBio.org as well. That’s another biodiversity data aggregator with a mission that differs slightly from GBIF’s, and a search portal that facilitates exploration in different ways, often complementing GBIF’s. We like, for example, that in iDigBio we can search for all specimens collected by any given collector, but that doesn’t work in GBIF. On the down side for iDigBio, we can’t provide links directly to different views or queries as we did above to help you explore our data in GBIF – any explorations in iDigBio’s portal have to start from scratch.
Specimen and associated data.
Getting our data into GBIF and iDigBio greatly increases the biodiversity data available to scientists. To help illustrate the general impact of our data contributions to the general knowledge of Texas' biodiversity, I queried GBIF for records within a rectangle that included all of Texas and, obviously, some adjacent areas. Restricting results to our own data for fishes, amphibians, and reptiles combined from that area produced 206,352 species occurrences, and when I broaden that to get data from the same area from all of the world's other data providers, the total jumps to 1,070,336. In other words, our own data comprise 24% of all of the world's data for those 3 groups in that area. Pretty impressive!
Since my taxonomic bias snuck into that example, let's do arthropods too. 13,513 of our own collection's records fall in that same polygon, and if I ask for everything the world provides for the same area, that's 458,948, meaning that the 1% of the insect collection Dr. Wild has digitized and published already comprises 3% of all the world's data for arthropods in that rectangle.
From a scientist's perspective, perhaps as remarkable as the sheer volume of data, is that they now come from both GBIF and iDigBio ready-to-use: One click at any stage of the process described above puts a file on my desktop that is formatted following global standards, ready for analysis in conjunction with anyone else’s data from GBIF (now serving 1,599,688,672 records) or any of the world’s other major biodiversity data aggregators, and easily linked to environmental data or most anything else I might want to throw into the mix. For perspective, only a decade ago, with a whole lot of effort (months probably) and personal help from the other curators, I might have been able to pull together the same data set, but it would have been substantially smaller and I'd then have to do a whole lot of data re-organizing, georeferencing and standardizing before I could use it. That's because each collection/discipline managed their data in their own unique ways, thus that huge investment in data compiling and cleaning made such research nearly impossible, until now.

Dung beetle specimen from Entomology Collection with tag. (Photo: Alex Wild)
That our combined data publishing efforts were almost to this point recently allowed me to obtain one of the new UT Stengl-Wyer Endowment grants. We hope to analyze the distributions of the Edwards Aquifer's blind catfishes, salamanders and invertebrates in the context of water quality and other environmental data. This grant will first accomplish the final step toward making all of that data available by adding our Aquatic Cave Invertebrate Collection (about 40,000 records) to what Entomology is already publishing to GBIF. Adding that broader taxonomic coverage and greatly increased number of occurrences to a combined analysis will hopefully go a long way toward furthering our understanding of what environmental parameters are important to the obscure Edwards Aquifer biota with its many endangered species.
Getting our data to this point has been anything but trivial. As Dr. Wild mentioned in his earlier post, challenges abound, such as lack of time and staff, poorly-labeled specimens, and huge backlogs. Additionally, curators are primarily biologists. Most lack training in informatics and general computer skills required to make digitization and global publication happen. However, the task of maintaining all this data publishing and updating it is now trivial. Our collections’ databases update automatically at pre-determined intervals (weekly for the fish data), so newly cataloged specimens are available for the world to use very quickly, and effortlessly on our part. We owe everything I’ve described in this blog in large part to the assistance provided to us by UT’s Texas Advanced Computing Center (TACC). The Fish Collection started working with the Data Management and Collections group over 15 years ago on the Fishes of Texas project, and before that they helped us move our data into the program we now use for all aspects of our own collection's management: Specify. Later, they did the same for all of the other Biodiversity Center's collections databases. They also set up our own Biodiversity Data publication system (IPT) to enable efficient publication to GBIF. We can't possibly afford our own staff to do all that Tomislav Urban of TACC has done for us. He is now functionally the primary database manager, maintainer and publisher for all of our biodiversity collections. Without him and TACC's more general technical support, our collections would likely have been left in the dust kicked up by the global rush to better document the planet's biodiversity.